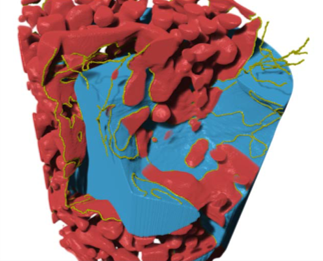
Visualize, explore, and quantify with best in class rendering
Dragonfly 3D World ZEISS edition is an advanced analysis software solution for 2D/3D data acquired by a variety of imaging technologies including X-ray microscopy and FIB-SEM. Using state-of-the-art volume rendering and access to advanced visualization techniques, Dragonfly 3D World ZEISS edition enables high-definition exploration into the details and properties of 3D datasets. Interactive inspection with color and opacity mapping means that in-depth analyses can always be visualized in meaningful ways and findings can be presented with easy-to-produce high-quality animated sequences.Multi-scale and multi-modal data visualization
With Dragonfly 3D World ZEISS edition, file formats such as .txm and .czi can be read and include system spatial coordinates allowing for accurate dataset registration used in Scout and Scan and correlative workflows. Combined with data handling for time-dependent, 4D data.Customize with Python scripting
Dragonfly 3D World ZEISS edition enables Python scripting - a powerful feature that allows users to customize their data processing and visualization needs. Users now have access to the data channels and properties within the interface creating a limitless and customizable post-processing environment. In addition, Dragonfly 3D World ZEISS edition uses Python with numpy, scipy and many other libraries standard with Anaconda distribution.Easy to use
Dragonfly 3D World ZEISS edition is easy to use. With minutes of experience, you can explore, navigate, and annotate your dataset. In a few hours, gain control of more advanced tools such as image processing, segmentation, object analysis, and movie making. Use our compact, topic-specific training videos to familiarize yourself with Dragonfly 3D World ZEISS edition in the shortest amount of time.
High-impact visualization
Image processing and segmentation
Correlative ready
|
Analyze
Import dataExport data |
Deep Learning module
Revolutionize your workflows: Powered by Google’s TensorFlow and Keras, Dragonfly 3D World ZEISS edition gives you the power to train, reuse, and repurpose existing models for advanced applications, and develop new neural networks.Dragonfly 3D World ZEISS edition's deep learning solution is bundled with pre-built and pre-trained neural networks, making it easy and fast to apply even for novices.
Vastly improve the quality of your images using these deep learning-based models. From denoising to super resolution, you will see your images in clearer detail than ever before.
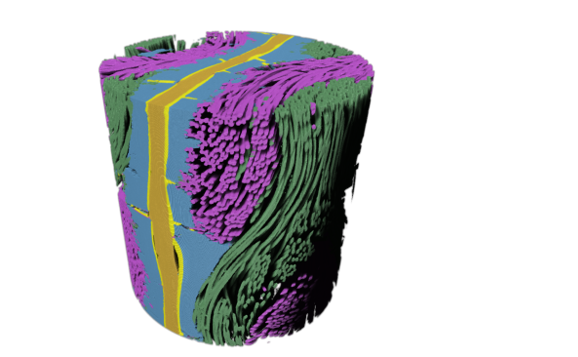
Deep Learning segmentation of polymer electrolyte fuel cell. |
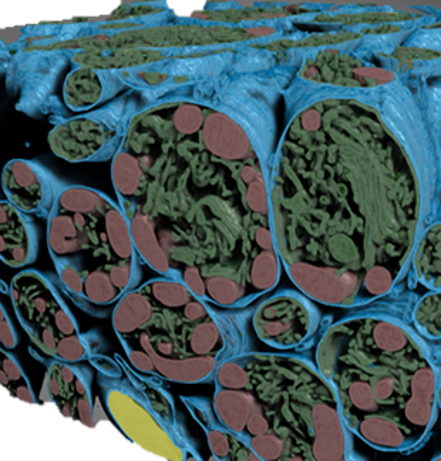
Deep Learning segmentation of mouse retina cell. |
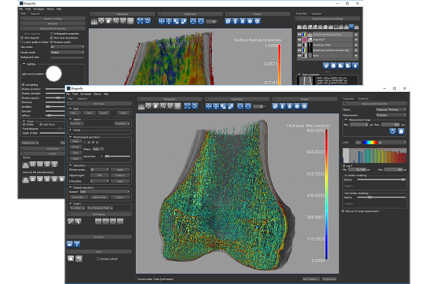
Bone Analysis Module
Analyze bone micro-architectures using semi-automated workflows. Designed for evaluating high-resolution micro-CT image data, the Bone Analysis Module provides 3D vector-based mappings of anisotropy magnitude and directionality and 3D scalar-based mappings of volume fraction. In addition, an automated separation of cortical and trabecular bone drives the calculation of common bone morphometric indices.Check out the technical note for more detail
|
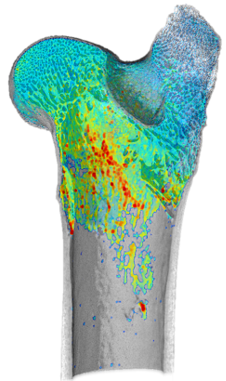
Volume thickness map of trabeculae |